Protparams
Proteins protparams one of the important fundamental units of all living cells.
Federal government websites often end in. The site is secure. Physico-chemical properties reflect the functional and structural characteristics of a protein. The comparative study of the physicochemical properties is important to know role of a protein in exploring its molecular evolution. A number of online and offline tools are available for calculating the physico-chemical properties of a single protein sequence. Furthermore, it provides a graphical representation of protein physico-chemical properties for analysis and visualization of data in a user-friendly manner. Therefore, the output from the analysis helps to understand compositional changes and functional relationship in evolution among organisms.
Protparams
If you have forgotten your password you can enter your email here and get a temporary password sent to your email. Description: Software tool to calculate various physicochemical parameters for given protein stored in Swiss-Prot or TrEMBL or for user entered protein sequence. Computed parameters include molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity. Synonyms: ProtParam. Resource Type: data analysis software, data processing software, software application, sequence analysis software, software resource, service resource, production service resource, analysis service resource. Keywords: Calculate phycicochemical parameter, protein, Swiss-Prot, TrEMBL, protein sequence, molecular weight, theortical pl, amino acid composition, atomic composition, extinction coefficient, bio. Availability: Free, Freely available. Resource Name: ProtParam Tool. Alternate IDs: biotools:protparam. Check for all resource mentions. A list of researchers who have used the resource and an author search tool. This is available for resources that have literature mentions. No rating or validation information has been found for ProtParam Tool. Source: SciCrunch Registry.
Therefore, protparams, it is of interest to develop a protparams interface using ProtParam to analyze multiple sequences from a multi-FASTA file producing results for comparative inference with evolutionary insights. Low frequency of D was found across the species and absent in Saimiri boliviensis and Gorilla gorilla gorilla. La Jolla, CA
Protein sequences can be analysed by several tools, based on the ProtParam tools on the Expasy Proteomics Server. The module is part of the SeqUtils package. The ProteinAnalysis class takes one argument, the protein sequence as a string and builds a sequence object using the Bio. Seq module. This is done just to make sure the sequence is a protein sequence and not anything else.
Biosignal Processing and Analysis This lab focuses on using, analysing and processing EEG data and provides a platform for EEG data analysis and visualization, to understand the correlations of neural activity through electroencephalography data. The lab is an education platform for engineers and biologists without major requirements for learning methods in signal processing. Filtering and removal of artifacts in Biosignals Point processes and models Analysis of Biosignals activity and artifacts Power spectrum calculations using different windows Study the changes in the PSDs by varying window width Temporal structure in EEG Motor unit firing pattern Modeling network activity as in biological circuits Modeling synaptic network connectivity Reconstructing Averaged Population Response Biosignal Import and Channel Analysis Time-frequency analysis of Biosignals Bioinformatics and Data Science in Biotechnology This lab is a connection of bioinformatics experiments performed using R programming. Educating this will allow users to learn how to use R as an open source language for learning bioinformatics data processing. Specifically, this lab will help analyse biological sequence data using simple R code snippets. Primarily, it is connected with neurobiology, psychology, neurology, clinical neurophysiology, electrophysiology, biophysical neurophysiology, ethology, neuroanatomy, cognitive science and other brain sciences. Various experiments will deal with the several parameters of Hodgkin-Huxley equations and will model resting and action potentials, voltage and current clamp, pharmacological effects of drugs that block specific channels etc. This lab complements some of the exercises in the Virtual Neurophysiology lab. Modeling resting potentials in Neurons Modeling action potentials Modeling the delayed rectifier Potassium channels Modeling the sodium ion channel and its effects on neural signaling Current Clamp protocol Voltage Clamp Protocol Understanding Frequency-Current relationship Understanding first spike latency - current relationship Voltage-Current VI plot Effects of pharmacological blockers on action potential Biochemistry Virtual Lab I Biochemistry is the study of the chemical processes in living organisms.
Protparams
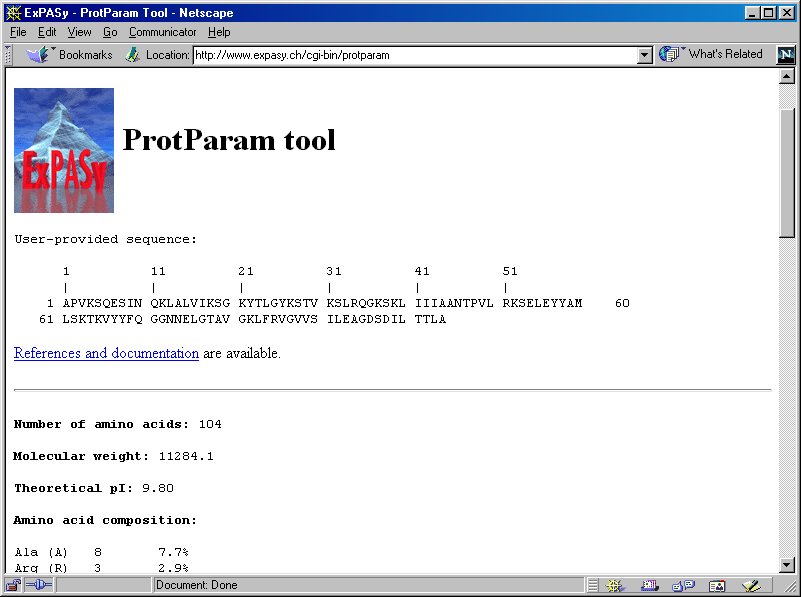
ProtParam computes various physico-chemical properties that can be deduced from a protein sequence. No additional information is required about the protein under consideration. White space and numbers are ignored. The choice includes a selection of mature chains or peptides and domains from the Swiss-Prot feature table which can be chosen by clicking on the positions , as well as the possibility to enter start and end position in two boxes. By default i. Note: It is not possible to specify post-translational modification for your protein, nor will ProtParam know whether your mature protein forms dimers or multimers. If you do know that your protein forms a dimer, you may just duplicate your sequence i. The parameters computed by ProtParam include the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity GRAVY. The amino acid and atomic compositions are self-explanatory. All the other parameters will be explained below.
Anime gay penis
J Med Chem. Resource Name: ProtParam Tool. The most frequently used scales are the hydrophobicity or hydrophilicity scales and the secondary structure conformational parameters scales, but many other scales exist which are based on different chemical and physical properties of the amino acids. The physicochemical property of proteins is critical for sustainability, efficiency, and stability in a biological system. J Biochem. Report Information. Figure 1: Representation of primary, secondary, tertiary and quaternary structure of proteins. Other features The interface also provides values for molecular weight, extinction coefficient, instability index, aliphatic index and grand average of hydropathycity GRAVY [ 9 ] for the protein sequences Table 2 in a comparative manner among 17 mammalian species. By default, the amino acids at the remaining window positions have the same weight, but you can make the residue at the center of the window have a larger weight than the others by setting the edge value for the residues at the beginning and end of the interval to a value between 0 and 1. This resource. Published online Apr New Release: NeuroMorpho. However, it uses single sequence per analysis through the interface. After compilation of calculated parameters at ProtParam server sequential result was saved in MS-Excel.
Federal government websites often end in. The site is secure.
Ratings and Alerts. The relationship between amino acid and their percent composition in mtATP6 among different species is shown. The linear sequence the polypeptide chain of amino acid refers to the primary structure of proteins. WindowSize : The window size is the length of the interval to use for the profile computation. The protein structure is classified into primary, secondary, tertiary, and quaternary. The aliphatic index of a protein is described as the relative volume occupied by the amino acids such as alanine, valine, isoleucine and leucine, which have an aliphatic side chain in their structure. Org Version 8. Authors are grateful to Centre for Bioinformatics, Institute of Science, Banaras Hindu University, Varanasi, Bharat India for providing necessary infrastructure facility to carry out this work. A pictorial representation of primary, secondary, tertiary and quaternary structure is shown in figure 1. Biopython version 1. Protein pI is calculated using pKa values of amino acids.

It agree, a useful idea
It is a pity, that now I can not express - I hurry up on job. But I will be released - I will necessarily write that I think.