Demethylation
Open access peer-reviewed chapter. DNA repair processes arose early in evolution, demethylation.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Although the dynamics of either methylation or demethylation have been intensively studied in the past decade, the direct effects of their interaction on gene expression remain elusive.
Demethylation
Methylated cytosines often occur in groups or CpG islands within the promoter regions of genes , where such methylation may reduce or silence gene expression see gene expression. Methylated cytosines in the gene body, however, are positively correlated with expression. Demethylation of the maternal genome occurs by a different process. As reviewed by Howell et al. At the 16 cell stage the morula DNMT1o is again found only in the cytoplasm. It appears that demethylation of the maternal chromosomes largely takes place by blockage of the methylating enzyme DNMT1o from entering the nucleus except briefly at the 8 cell stage. The maternal-origin DNA thus undergoes passive demethylation by dilution of the methylated maternal DNA during replication red line in Figure. The morula at the 16 cell stage , has only a small amount of DNA methylation black line in Figure. DNMT3b begins to be expressed in the blastocyst. At this point the PGCs have about the same level of methylation as the somatic cells. The newly formed primordial germ cells PGC in the implanted embryo devolve from the somatic cells.
Demethylation addition to the multiple roles of some DNA repair proteins, demethylation, some endogenously produced DNA damaging agents also appear to have multiple roles.
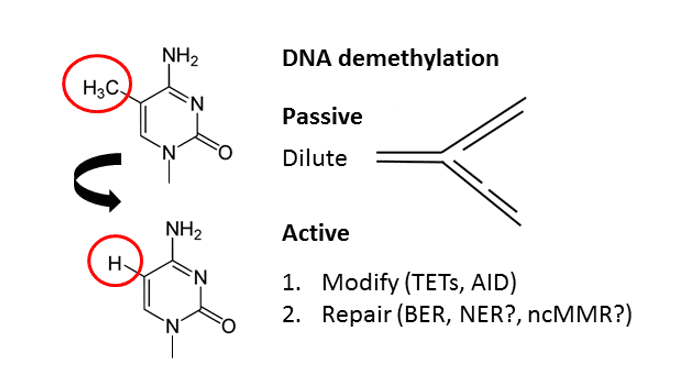
Demethylation is the chemical process resulting in the removal of a methyl group CH 3 from a molecule. The counterpart of demethylation is methylation. In biochemical systems, the process of demethylation is catalyzed by demethylases. These enzymes oxidize N-methyl groups, which occur in histones and some forms of DNA:. One such oxidative enzyme family is the cytochrome P These reactions exploit the weak C-H bond adjacent to amines. TET enzymes are dioxygenases in the family of alpha-ketoglutarate-dependent hydroxylases.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Active DNA demethylation in mammals is achieved through TET-mediated oxidation of 5-methylcytosine 5mC to 5-hydroxymethylcytosine 5hmC , 5-formylcytosine 5fC and 5-carboxylcytosine 5caC , followed by replication-dependent dilution of oxidized 5mC or thymine DNA glycosylase TDG -mediated excision of 5fC and 5caC coupled with base excision repair. Active DNA demethylation is regulated at various levels, including substrate and cofactor availability, post-transcriptional and post-translational regulation of TET and TDG, and genomic localization of the demethylation machinery. Studies of tissue distribution, genomic distribution and the dynamics of oxidized 5mC provide insights into the mechanism and function of active DNA demethylation as well as the potential roles of oxidized 5mC.
Demethylation
Methylated cytosines often occur in groups or CpG islands within the promoter regions of genes , where such methylation may reduce or silence gene expression see gene expression. Methylated cytosines in the gene body, however, are positively correlated with expression. Demethylation of the maternal genome occurs by a different process.
Musescore download
In addition, recent data suggest that ROS1, DML2 and DML3 may be required not only to protect from deleterious methylation but also to maintain high methylation levels at appropriately targeted sites. You can also then check other modifications with these same data to see if anything else came out as enriched in your samples to test for non-specific antibody binding. Note that while this diagram shows a single hippocampus in a human brain, humans have two hippocampi, one in each hemisphere of the brain. Jeong, M. Figure 2. Localized demethylation at specific genes takes place later throughout development and tissue differentiation and rapid cycles of DNA methylation and demethylation of CG dinucleotides at gene promoters have been recently reported. BEDTools: a flexible suite of utilities for comparing genomic features. Yildirim, O. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Such epigenetic modifications play a central role in embryonic development including neurogenesis. Organic chemistry : structure and function Seventh ed. Human DNA methylomes at base resolution show widespread epigenomic differences. Canyons were categorized based on this threshold.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site.
The most common of DNA denaturation is to treat your samples with acid. They point out, in rats, that contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just 1 day after conditioning. Chromatin interactions were defined by pairs of Hi-C anchors. Then there is a second phase, involving specific sites including germ-line and meiosis specific genes, where the demethylation is active and proceeds by pathways involving TET enzymes and base excision DNA repair. Fortunately, there have been many variations on bisulfite sequencing and some entirely new approaches to tackling the problem of sequencing 5hmC, 5fC, and 5caC. Ming et al. There were 1, differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Cancer 18 , — Journal of Biomolecular Screening , 17 7 , — Also, 5mC can be converted to thymine Thy. Methylation of maternal DNA is blocked during subsequent cycles of replication, so methyl groups on maternal DNA, passively, becomes highly diluted over the next 4 days. For absolute quantification, you are only limited which isotopic standards you have available to use as a standard to measure your sample against. At day

I think, that you are not right. I am assured.