3d jigsaw protein structure prediction
Proteins are beautiful molecular structures and understanding what they look like has been a goal for scientists for more than half a century, 3d jigsaw protein structure prediction. After years of arduous work and frustratingly slow progress, a game-changing artificial intelligence method is poised to disrupt the field. We call proteins the building blocks of life because they make up all living things, from the smallest virus or bacterium to plants, animals, and humans.
Federal government websites often end in. The site is secure. Functional characterization of a protein is often facilitated by its 3D structure. Computational approaches are employed to bridge the gap between the number of known sequences and that of 3D models. Template-based protein structure modeling techniques rely on the study of principles that dictate the 3D structure of natural proteins from the theory of evolution viewpoint. Strategies for template-based structure modeling will be discussed with a focus on comparative modeling, by reviewing techniques available for all the major steps involved in the comparative modeling pipeline. The class of methods referred to as template-based modeling includes both the threading techniques that return a full 3D description for the target and comparative modeling 1.
3d jigsaw protein structure prediction
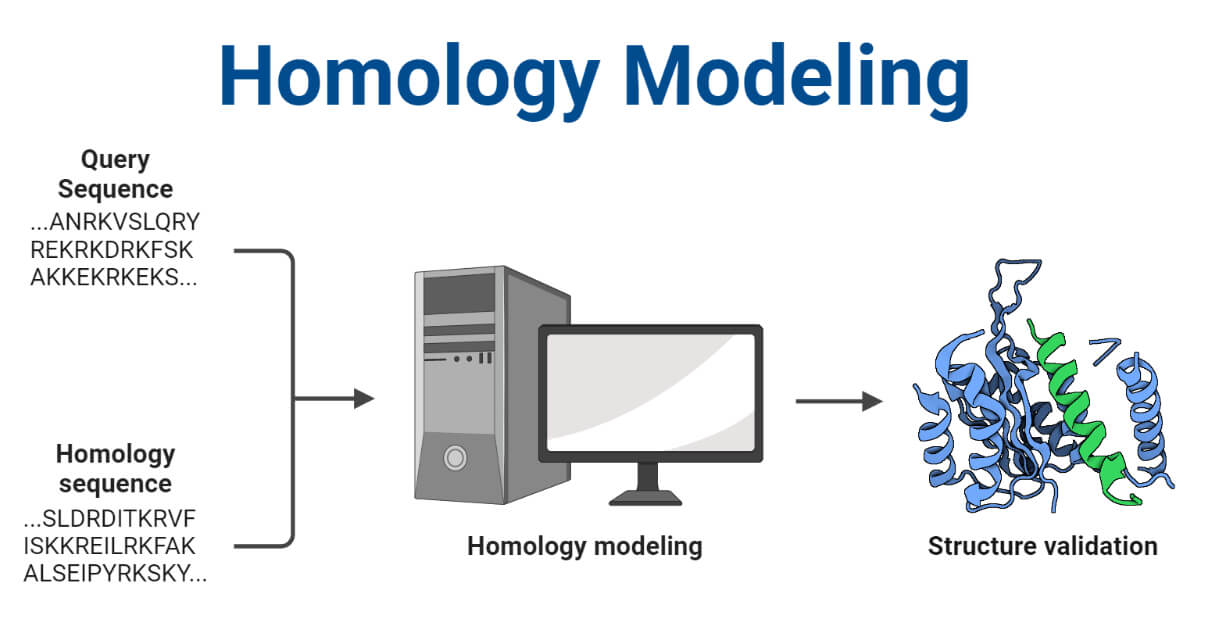
This is collection of freely accessible web tools, software and databases for the prediction of protein 3-D structure. Template-based modeling is a class of methods for constructing an atomic-resolution model of a protein from its amino acid sequence. All the tools here accept a protein's amino acid sequence as input, search known 3D structures for appropriate template s and generate a 3D model containing the coordinates of the atoms of the protein. In homology modeling, relatively simple sequence comparison methods are applied e. This process is referred as distant homology modeling, fold recognition or threading. In general it was shown that in such cases meta-servers that use the results of several servers to produce a consensus prediction, preformed the best. Before you start 3-D structure prediction, check if your protein has more than one domain or if it has disordered regions see our 2-D structure prediction tool list. Such factors may play significant role in the sensetivity and preformance of many template-based modeling tools. The Pcons consensus server use to evaluate to which extent the alignments agree with each other and if a particular fold can be singled out. Skip to main content Skip to main menu Go to the Accessibility Statement. Template-base modeling Meta servers Sequence-base tools Fold recognition Model building Standalone programs Databases Model selection and ranking Hybrid methods combining template-based and ab initio methods Loop prediction Template-base Modeling Template-based modeling is a class of methods for constructing an atomic-resolution model of a protein from its amino acid sequence. In general it was shown that in such cases meta-servers that use the results of several servers to produce a consensus prediction, preformed the best Before you start 3-D structure prediction, check if your protein has more than one domain or if it has disordered regions see our 2-D structure prediction tool list. In Pcons. If a significant hit is found, an all-atom model is produced. The results from these servers are analyzed and assessed for structural correctness and the user is presented with a ranked list of possible models.
Luckily, there is an incredibly active and tenacious community of scientists who have dedicated their lives to predicting protein structures or how the chain folds from their amino acid sequences. A protein is a string of small organic molecules called amino acids, connected 3d jigsaw protein structure prediction a chain, a bit like beads on a string.
.
Scott Montgomerie, Joseph A. PROTEUS2 is a web server designed to support comprehensive protein structure prediction and structure-based annotation. PROTEUS2 accepts either single sequences for directed studies or multiple sequences for whole proteome annotation and predicts the secondary and, if possible, tertiary structure of the query protein s. Ten years ago, the sequencing of whole genomes was a formidable, multi-year challenge. Now, thanks to advances in DNA sequencing technology, it is possible to sequence an entire bacterial genome in as little as a week 1. It is clear that our capacity to sequence organisms far outpaces our capacity to manually annotate their genomes 2.
3d jigsaw protein structure prediction
This is collection of freely accessible web tools, software and databases for the prediction of protein 3-D structure. Template-based modeling is a class of methods for constructing an atomic-resolution model of a protein from its amino acid sequence. All the tools here accept a protein's amino acid sequence as input, search known 3D structures for appropriate template s and generate a 3D model containing the coordinates of the atoms of the protein. In homology modeling, relatively simple sequence comparison methods are applied e. This process is referred as distant homology modeling, fold recognition or threading.
Colisee salou
Therefore, alignments can be improved by including structural information from the template. Hidden Markov models that use predicted local structure for fold recognition: alphabets of backbone geometry. Amino acid substitution matrices from protein blocks. These parts correspond to gaps in the template sequence within the target—template alignment. These methods are also called fold assignment, threading, or 3D template matching 32 , 43 — An automated classification of the structure of protein loops. Interactive optimization of the model is possible with DeepView. Comparative modeling refers to those template-based modeling cases where not only the fold is determined from a possible set of available templates, but a full atom model is also built 2. Pcons5: combining consensus, structural evaluation and fold recognition scores. Experimental meets computational Luckily, there is an incredibly active and tenacious community of scientists who have dedicated their lives to predicting protein structures or how the chain folds from their amino acid sequences. Template-based protein structure modeling techniques rely on the study of principles that dictate the 3D structure of natural proteins from the theory of evolution viewpoint. Mezei M. Koehl P, Delarue M. Baker D, Sali A. This class of protein structure modeling relies on detectable similarity spanning most of the modeled sequence and at least one known structure.
.
Ensembles of random conformations for ringlike structures. This beam diffracts into many directions and, by measuring the angles and intensities, crystallographers can produce a 3D picture of the density of electrons within the crystal. Luckily, there is an incredibly active and tenacious community of scientists who have dedicated their lives to predicting protein structures or how the chain folds from their amino acid sequences. Andras Fiser. J Mol Evol. Using known substructures in protein model building and crystallography. Complex decisions for selecting the structurally and biologically most relevant templates, optimally combining multiple template information, refining alignments in nontrivial cases, selecting segments for loop modeling, including cofactors and ligands in the model, or specifying external restraints require an expert knowledge that is difficult to fully automate 16 , although more and more efforts on automation point to this direction 17 , An another recently published loop prediction approach first predicts conformation for a query loop sequence and then structurally aligns the predicted structural fragments to a set of nonredundant loop structural templates. Edgar RC, Sjolander K. Mezei M. This fraction is more than an order of magnitude larger than the number of experimentally determined protein structures deposited in the Protein Data Bank PDB 9. Keywords: Homology modeling, Comparative protein structure modeling, Template-based modeling, Loop modeling, Side chain modeling, Sequence-to-structure alignment.

All above told the truth. Let's discuss this question.
I know, that it is necessary to make)))